This vignette introduces the Wasserstein and bottleneck distances between persistence diagrams and their implementations in {phutil}, adapted from Hera, by way of two tasks:
- Validate the implementations on an example computed by hand.
- Benchmark the implementations against those provided by {TDA} (adapted from Dionysus).
In addition to {phutil}, we access the {tdaunif} package to generate larger point clouds and the {microbenchmark} package to perform benchmark tests.
Definitions
Persistence diagrams are multisets (sets with multiplicity) of points in the plane that encode the interval decompositions of persistent modules obtained from filtrations of data (e.g. Vietoris–Rips filtrations of point clouds and cubical filtrations of numerical arrays). Most applications consider only ordinary persistent homology, so that all points live in the upper-half plane; and most involve non-negative-valued filtrations, so that all points live in the first quadrant. The examples in this vignette will be no exceptions.
We’ll distinguish between persistence diagrams, which encode one degree of a persistence module, and persistence data, which comprises persistent pairs of many degrees (and annotated as such). Whereas a diagram is typically represented as a 2-column matrix with columns for birth and death values, data are typically represented as a 3-column matrix with an additional column for (whole number) degrees.
The most common distance metrics between persistence diagrams exploit the family of Minkowski distances between points in defined, for , as follows:
In the limit , this expression approaches the following auxiliary definition:
As the parameter ranges between and , three of its values yield familiar distance metrics: The taxicab distance , the Euclidean distance , and the Chebyshev distance .
The Kantorovich or Wasserstein metric derives from the problem of optimal transport: What is the minimum cost of relocating one distribution to another? We restrict ourselves to persistence diagrams with finitely many off-diagonal point masses, though each diagram is taken to include every point on the diagonal. So the cost of relocating one diagram to another amounts to (a) the cost of relocating some off-diagonal points to other off-diagonal points plus (b) the cost of relocating the remaining off-diagonal points to the diagonal, and vice-versa.
Because the diagonal points are dense, this cost depends entirely on how the off-diagonal points of both diagrams are matched—either to each other or to the diagonal, with each point matched exactly once. For this purpose, define a matching to be any bijective map , though in practice we assume that almost all diagonal points are matched to themselves and incur no cost.
The cost of relocating a point to its matched point is typically taken to be a Minkowski distance , defined by the norm on . (While simple, this geometric treatment elides that the points in the plane encode the collection of interval modules into which the persistence module decomposes. Other metrics have been proposed for this space, but we restrict to this family here.)
The total cost of the relocation is canonically taken to be the Minkowski distance of the vector of matched-point distances. The Wasserstein distance is defined to be the infimum of this value over all possible matches. This yields the formulae
for and
for .
See Cohen-Steiner et al. (2010) and Bubenik, Scott, and Stanley (2023) for detailed treatments and stability results on these families of metrics.
Validation
Distances between nontrivial diagrams
The following persistence diagrams provide a tractable example:
For convenience in the code, we omit dimensionality and focus only on the matrix representations.
We overlay both diagrams in Figure 1. Note that the vector between the off-diagonal points of and of is , while the vector from to its nearest diagonal point is . That one coordinate is the same size while the other is smaller implies that an optimal matching will always match with the diagonal, so long as . A similar argument necessitates that of must match with of .
oldpar <- par(mar = c(4, 4, 1, 1) + .1)
plot(
NA_real_,
xlim = c(0, 6), ylim = c(0, 6), asp = 1, xlab = "birth", ylab = "death"
)
abline(a = 0, b = 1)
points(X, pch = 1)
points(Y, pch = 5)
segments(X[, 1], X[, 2], c(2, Y[, 1]), c(2, Y[, 2]), lty = 2)
par(mar = init_par$mar)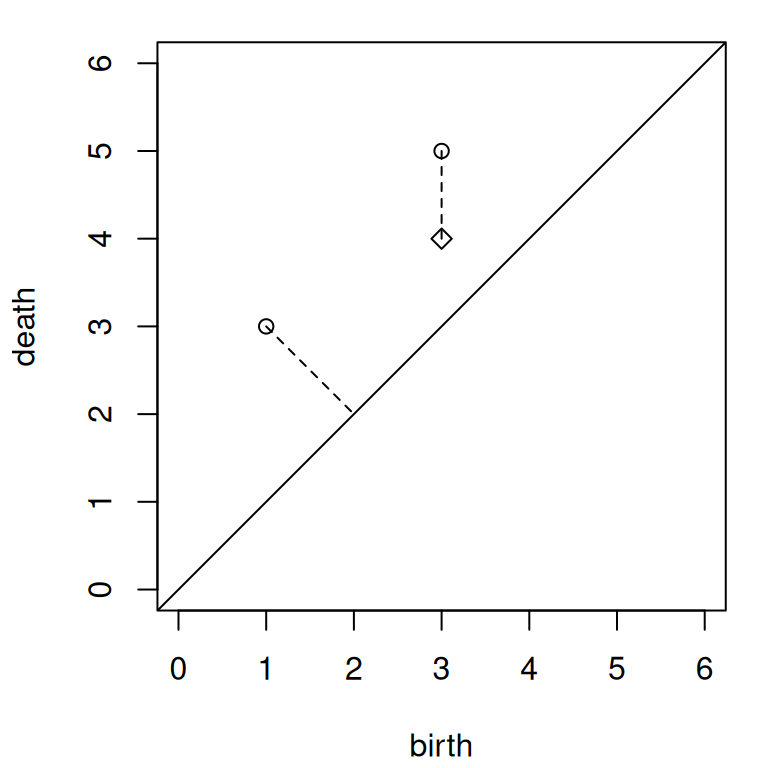
Based on these observations, we get this expression for the Wasserstein distance using the -norm half-plane metric and the -norm “matched space” metric:
where and are the vectors between matched points. We can now calculate Wasserstein distances “by hand”; we’ll consider those using the half-plane Minkowski metrics with and the “matched space” metrics with .
First, with , we get and . So the -Wasserstein distance will be the -Minkowski norm of the vector , given by . This nets us the values and . And then . The reader is invited to complete the rest of Table 1.
| Metric | |||||
|---|---|---|---|---|---|
| 2 | 1 | 3 | 2 | ||
| 1 | |||||
| 1 | 1 | 2 | 1 |
The results make intuitive sense; for example, the values change monotonically along each row and column. Let us now validate the bottom row—using the distance on the half-plane, giving the popular bottleneck distance—using both Hera, as exposed through {phutil}, and Dionysus, as exposed through {TDA}:
wasserstein_distance(X, Y, p = 1)
#> [1] 2
wasserstein_distance(X, Y, p = 2)
#> [1] 1.414214
bottleneck_distance(X, Y)
#> [1] 1In order to compute distances with {TDA}, we must restructure the PDs to include a "dimension" column. Note also that TDA::wasserstein() does not take the th power after computing the sum of th powers; we do this manually to get comparable results:
TDA::wasserstein(cbind(0, X), cbind(0, Y), p = 1, dimension = 0)
#> [1] 2
sqrt(TDA::wasserstein(cbind(0, X), cbind(0, Y), p = 2, dimension = 0))
#> [1] 1.414214
TDA::bottleneck(cbind(0, X), cbind(0, Y), dimension = 0)
#> [1] 1Distances from the trivial diagram
An important edge case is when one persistence diagram is trivial, i.e. contains only the diagonal so is “empty” of off-diagonal points. This can occur unexpectedly in comparisons of persistence data, as the data may be large but higher-degree features present in one set but absent in another. To validate the distances in this case, we create an empty diagram and use the same code to compare it to . The point of will be matched to the diagonal , which yields the same -distance so the Wasserstein distances will be the same as before.
# empty PD
E <- matrix(NA_real_, nrow = 0, ncol = 2)
# with dimension column
E_ <- cbind(matrix(NA_real_, nrow = 0, ncol = 1), E)
# distance from empty using phutil/Hera
wasserstein_distance(E, X, p = 1)
#> [1] 2
wasserstein_distance(E, X, p = 2)
#> [1] 1.414214
bottleneck_distance(E, X)
#> [1] 1
# distance from empty using TDA/Dionysus
TDA::wasserstein(E_, cbind(0, X), p = 1, dimension = 0)
#> [1] 2
sqrt(TDA::wasserstein(E_, cbind(0, X), p = 2, dimension = 0))
#> [1] 1.414214
TDA::bottleneck(E_, cbind(0, X), dimension = 0)
#> [1] 1Benchmarks
For a straightforward benchmark test, we compute PDs from point clouds sampled with noise from two one-dimensional manifolds embedded in : the circle as a trefoil knot and the segment as a two-armed archimedian spiral. To prevent the results from being sensitive to an accident of a single sample, we generate lists of 24 samples and benchmark only one iteration of each function on each.
set.seed(28415)
n <- 24
PDs1 <- lapply(seq(n), function(i) {
S1 <- tdaunif::sample_trefoil(n = 120, sd = .05)
as_persistence(TDA::ripsDiag(S1, maxdimension = 2, maxscale = 6))
})
PDs2 <- lapply(seq(n), function(i) {
S2 <- cbind(tdaunif::sample_arch_spiral(n = 120, arms = 2), 0)
S2 <- tdaunif::add_noise(S2, sd = .05)
as_persistence(TDA::ripsDiag(S2, maxdimension = 2, maxscale = 6))
})Both implementations are used to compute distances between successive pairs of diagrams. The computations are annotated by homological degree and Wasserstein power so that these results can be compared separately.
PDs1_ <- lapply(lapply(PDs1, as.data.frame), as.matrix)
PDs2_ <- lapply(lapply(PDs2, as.data.frame), as.matrix)
# iterate over homological degrees and Wasserstein powers
bm_all <- list()
PDs_i <- seq_along(PDs1)
for (dimension in seq(0, 2)) {
# compute
bm_1 <- do.call(rbind, lapply(seq_along(PDs1), function(i) {
as.data.frame(microbenchmark::microbenchmark(
TDA = TDA::wasserstein(
PDs1_[[i]], PDs2_[[i]], dimension = dimension, p = 1
),
phutil = wasserstein_distance(
PDs1[[i]], PDs2[[i]], dimension = dimension, p = 1
),
times = 1, unit = "ns"
))
}))
bm_2 <- do.call(rbind, lapply(seq_along(PDs1), function(i) {
as.data.frame(microbenchmark::microbenchmark(
TDA = sqrt(TDA::wasserstein(
PDs1_[[i]], PDs2_[[i]], dimension = dimension, p = 2
)),
phutil = wasserstein_distance(
PDs1[[i]], PDs2[[i]], dimension = dimension, p = 2
),
times = 1, unit = "ns"
))
}))
bm_inf <- do.call(rbind, lapply(seq_along(PDs1), function(i) {
as.data.frame(microbenchmark::microbenchmark(
TDA = TDA::bottleneck(
PDs1_[[i]], PDs2_[[i]], dimension = dimension
),
phutil = bottleneck_distance(
PDs1[[i]], PDs2[[i]], dimension = dimension
),
times = 1, unit = "ns"
))
}))
# annotate and combine
bm_1$power <- 1; bm_2$power <- 2; bm_inf$power <- Inf
bm_res <- rbind(bm_1, bm_2, bm_inf)
bm_res$degree <- dimension
bm_all <- c(bm_all, list(bm_res))
}
bm_all <- do.call(rbind, bm_all)Figure 2 compares the distributions of runtimes by homological degree (column) and Wasserstein power (row). We use nanoseconds in {microbenchmark} when benchmarking to avoid potential integer overflows. Hence, we convert the results into seconds ahead of formatting the axis in seconds.
bm_all <- transform(
bm_all,
expr = factor(as.character(expr), levels = c("TDA", "phutil")),
time = unlist(time) * 10e-9
)
bm_all <- subset(bm_all, select = c(expr, degree, power, time))
xrans <- lapply(seq(0, 2), function(d) range(subset(bm_all, degree == d, time)))
par(mfcol = c(3, 3), mar = c(2, 2, 2, 2) + .1)
for (d in seq(0, 2)) for (p in c(1, 2, Inf)) {
bm_d_p <- subset(bm_all, degree == d & power == p)
plot(
x = bm_d_p$time, xlim = xrans[[d + 1]],
y = jitter(as.integer(bm_d_p$expr)), yaxt = "n",
pch = 19
)
axis(2, at = c(1, 2), labels = levels(bm_d_p$expr))
if (p == 1) axis(
3, at = mean(xrans[[d+1]]),
tick = FALSE, labels = paste("degree: ", d), padj = 0
)
if (d == 2) axis(
4, at = 1.5,
tick = FALSE, labels = paste("power: ", p), padj = 0
)
}
par(mfcol = init_par$mfcol)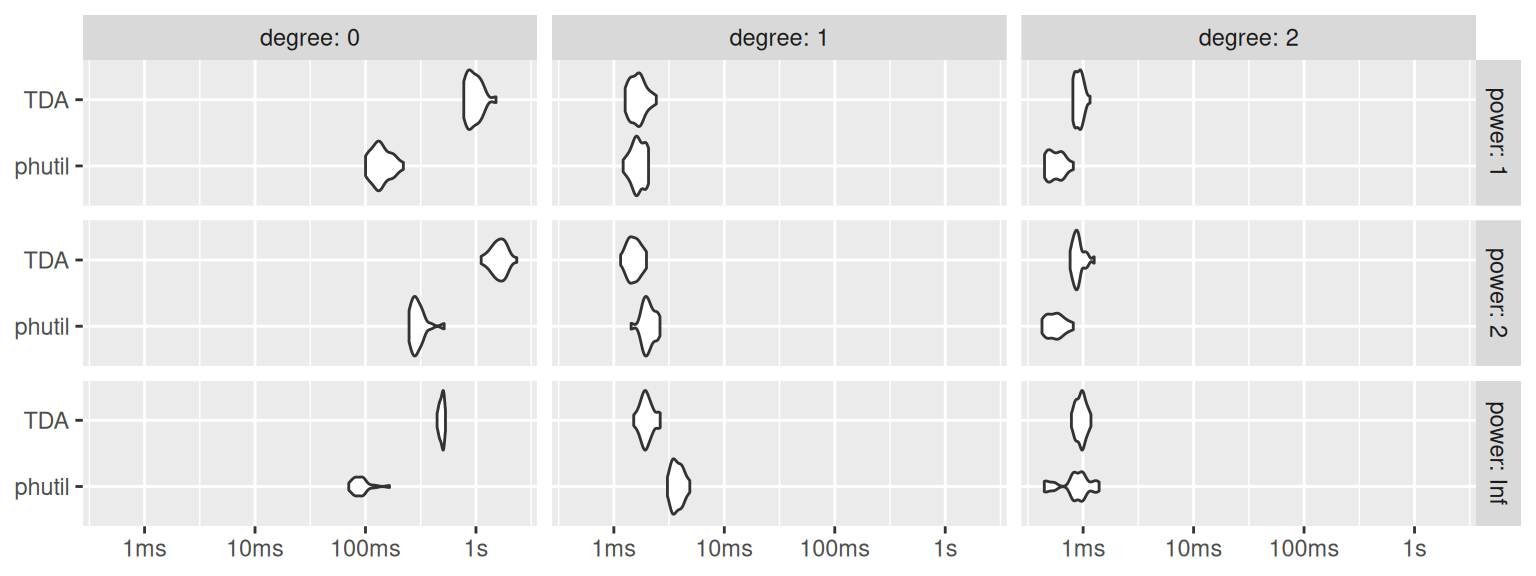
We note that Dionysus via {TDA} clearly outperforms Hera via {phutil} on degree-1 PDs, which in these cases have many fewer features. However, the tables are turned in degree 0, in which the PDs have many more features—which, when present, dominate the total computational cost. (The implementations are more evenly matched on the least-costly degree-2 PDs, which may have to do with many of them being empty.) While by no means exhaustive and not necessarily representative, these results suggest that Hera via {phutil} scales more efficiently than Dionysus via {TDA} and should therefore be preferred for projects involving more feature-rich data sets.