Visualize persistence data in a (flat, diagonal, or landscape) persistence diagram.
stat_persistence(
mapping = NULL,
data = NULL,
geom = "point",
position = "identity",
filtration = "Rips",
diameter_max = NULL,
radius_max = NULL,
dimension_max = 1L,
field_order = 2L,
engine = NULL,
order_by = c("persistence", "start"),
decreasing = FALSE,
diagram = "diagonal",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
...
)
geom_fundamental_box(
mapping = NULL,
data = NULL,
stat = "identity",
position = "identity",
diagram = "diagonal",
t = NULL,
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
...
)Arguments
- mapping
Set of aesthetic mappings created by
aes(). If specified andinherit.aes = TRUE(the default), it is combined with the default mapping at the top level of the plot. You must supplymappingif there is no plot mapping.- data
The data to be displayed in this layer. There are three options:
If
NULL, the default, the data is inherited from the plot data as specified in the call toggplot().A
data.frame, or other object, will override the plot data. All objects will be fortified to produce a data frame. Seefortify()for which variables will be created.A
functionwill be called with a single argument, the plot data. The return value must be adata.frame, and will be used as the layer data. Afunctioncan be created from aformula(e.g.~ head(.x, 10)).- geom
The geometric object to use to display the data, either as a
ggprotoGeomsubclass or as a string naming the geom stripped of thegeom_prefix (e.g."point"rather than"geom_point")- position
Position adjustment, either as a string naming the adjustment (e.g.
"jitter"to useposition_jitter), or the result of a call to a position adjustment function. Use the latter if you need to change the settings of the adjustment.- filtration
The type of filtration from which to compute persistent homology; one of
"Rips","Vietoris"(equivalent) or"alpha".- diameter_max, radius_max
Maximum diameter or radius for the simplicial filtration. Both default to
NULL, in which case the complete filtration is constructed.- dimension_max
Maximum dimension of the simplicial filtration.
- field_order
(Prime) order of the field over which to compute persistent homology.
- engine
The computational engine to use (see 'Details'). Reasonable defaults are chosen based on
filtration.- order_by
A character vector of required or computed variables (
"start","end","part", and/or"persistence") by which the features should be ordered (withingroup); defaults toc("persistence", "start"). This will most notably impact the appearance of barcodes.- decreasing
Logical; whether to sort features by decreasing values of
order_by(again, withingroup).- diagram
One of
"flat","diagonal", or"landscape"; the orientation for the diagram should take.- na.rm
Logical: if
FALSE, the default,NAlodes are not included; ifTRUE,NAlodes constitute a separate category, plotted in grey (regardless of the color scheme).- show.legend
logical. Should this layer be included in the legends?
NA, the default, includes if any aesthetics are mapped.FALSEnever includes, andTRUEalways includes. It can also be a named logical vector to finely select the aesthetics to display.- inherit.aes
If
FALSE, overrides the default aesthetics, rather than combining with them. This is most useful for helper functions that define both data and aesthetics and shouldn't inherit behaviour from the default plot specification, e.g.borders().- ...
Additional arguments passed to
ggplot2::layer().- stat
The statistical transformation to use on the data for this layer, either as a
ggprotoGeomsubclass or as a string naming the stat stripped of thestat_prefix (e.g."count"rather than"stat_count")- t
A numeric vector of time points at which to place fundamental boxes.
Details
Persistence diagrams are scatterplots of persistence data.
Persistence data
Persistence data encode the values of an underlying parameter \(\epsilon\) at which topological features appear ("birth") and disappear ("death"). The difference between the birth and the death of a feature is called its persistence. Whereas topological features may be of different dimensions, persistence data sets usually also include the dimension of each feature.
ggtda expects persistence data to have at least three columns: birth, death, and dimension.
Persistence diagrams
Persistence diagrams recognize extended persistence data, with negative birth/death values arising from the relative part of the filtration.
The original persistence diagrams plotted persistence against birth in what we call "flat" diagrams, but most plot death against birth in "diagonal" diagrams, often with a diagonal line indicating zero persistence.
The geom_fundamental_box() layer renders fundamental boxes at specified
time points (Chung & Lawson, 2020).
Aesthetics
stat_persistence() understands the following aesthetics (required aesthetics are in bold):
startordatasetendordatasetgroup
geom_fundamental_box() understands the following aesthetics (required aesthetics are in bold):
alphacolourfillgrouplinetypelinewidth
Learn more about setting these aesthetics in vignette("ggplot2-specs", package = "ggplot2").
Computed variables
stat_persistence calculates the following variables that can be accessed with delayed evaluation.
after_stat(start)
birth value of each feature (from 'dataset' aesthetic).after_stat(end)
death value of each feature (from 'dataset' aesthetic).after_stat(dimension)
integer feature dimension (from 'dataset' aesthetic).after_stat(group)
interaction of existing 'group', dataset ID, and 'dimension'.after_stat(id)
character feature identifier (across 'group').after_stat(part)
whether features belong to ordinary, relative, or extended homology.after_stat(persistence)
differences between birth and death values of features.
References
H Edelsbrunner, D Letscher, and A Zomorodian (2000) Topological persistence and simplification. Proceedings 41st Annual Symposium on Foundations of Computer Science, 454--463. doi:10.1109/SFCS.2000.892133
H Edelsbrunner and D Morozov (2012) Persistent Homology: Theory and Practice. European Congress of Mathematics, 31--50. doi:10.4171/120
Y-M Chung and A Lawson (2020) Persistence Curves: A Canonical Framework for Summarizing Persistence Diagrams. https://arxiv.org/abs/1904.07768
See also
ggplot2::layer() for additional arguments.
Examples
# toy example
toy.data <- data.frame(
birth = c(0, 0, 1, 2, 1.5),
death = c(5, 3, 5, 3, 6),
dim = c("0", "0", "2", "1", "1")
)
# diagonal persistence diagram, coding persistence to transparency
ggplot(toy.data,
aes(start = birth, end = death, colour = dim, shape = dim)) +
theme_persist() +
coord_equal() +
stat_persistence(aes(alpha = after_stat(persistence)),
diagram = "diagonal", size = 3) +
geom_abline(intercept = 0, slope = 1) +
lims(x = c(0, 6), y = c(0, 6)) +
guides(alpha = "none")
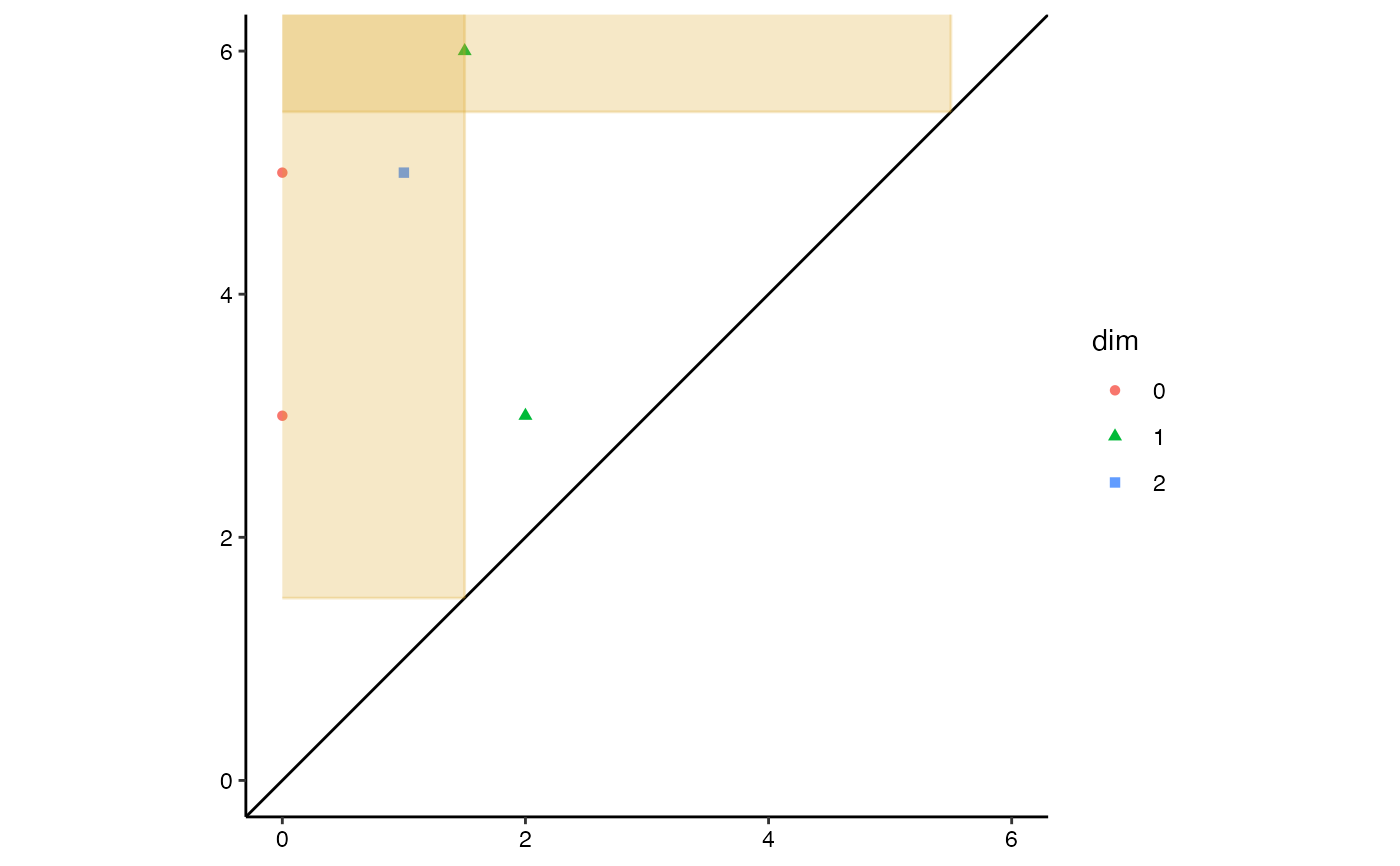
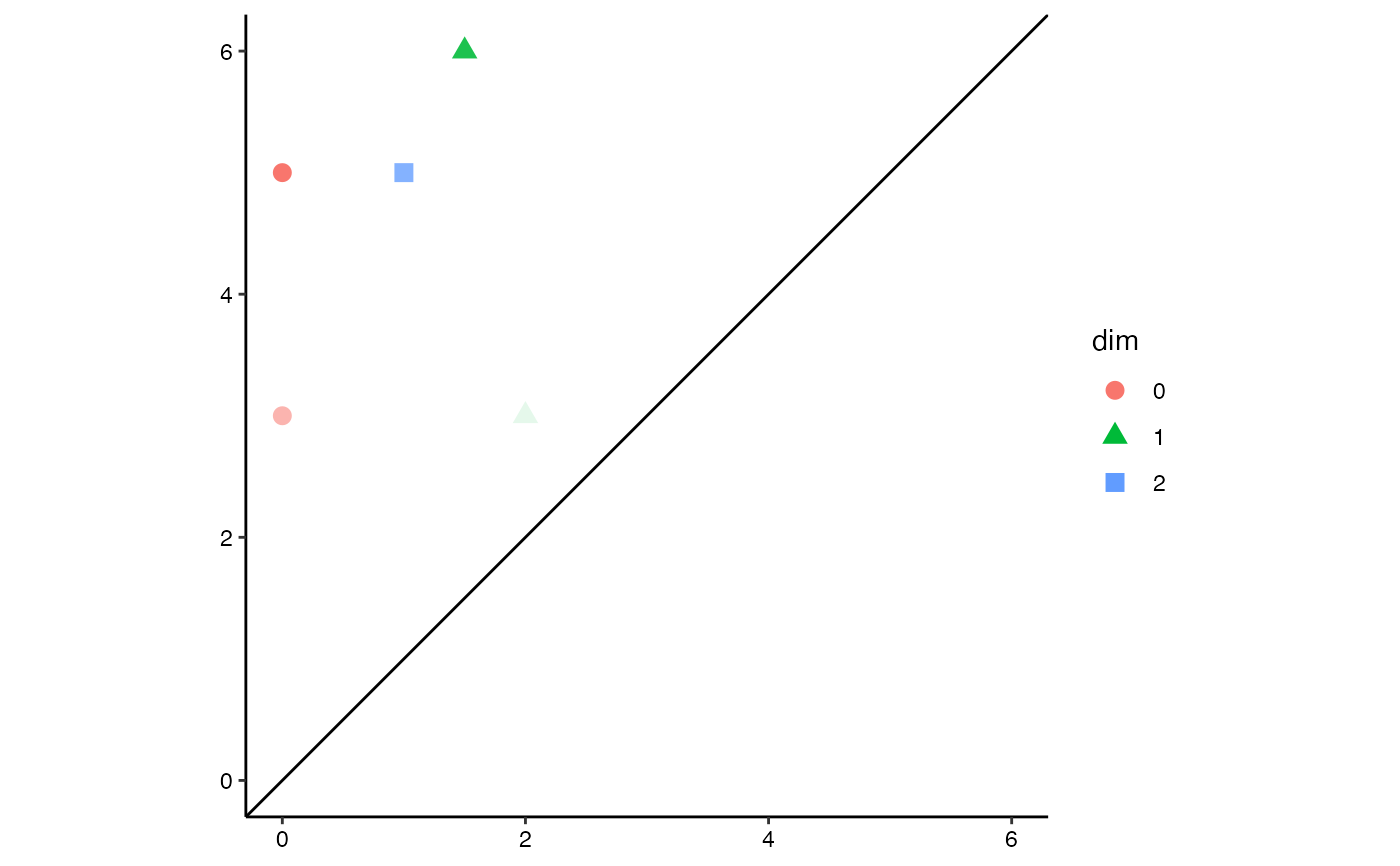 # diagonal persistence diagram with fundamental boxes
ggplot(toy.data,
aes(start = birth, end = death, colour = dim, shape = dim)) +
theme_persist() +
coord_equal() +
stat_persistence() +
geom_abline(intercept = 0, slope = 1) +
geom_fundamental_box(t = c(1.5, 5.5),
color = "goldenrod", fill = "goldenrod") +
lims(x = c(0, 6), y = c(0, 6)) +
guides(alpha = "none")
# diagonal persistence diagram with fundamental boxes
ggplot(toy.data,
aes(start = birth, end = death, colour = dim, shape = dim)) +
theme_persist() +
coord_equal() +
stat_persistence() +
geom_abline(intercept = 0, slope = 1) +
geom_fundamental_box(t = c(1.5, 5.5),
color = "goldenrod", fill = "goldenrod") +
lims(x = c(0, 6), y = c(0, 6)) +
guides(alpha = "none")
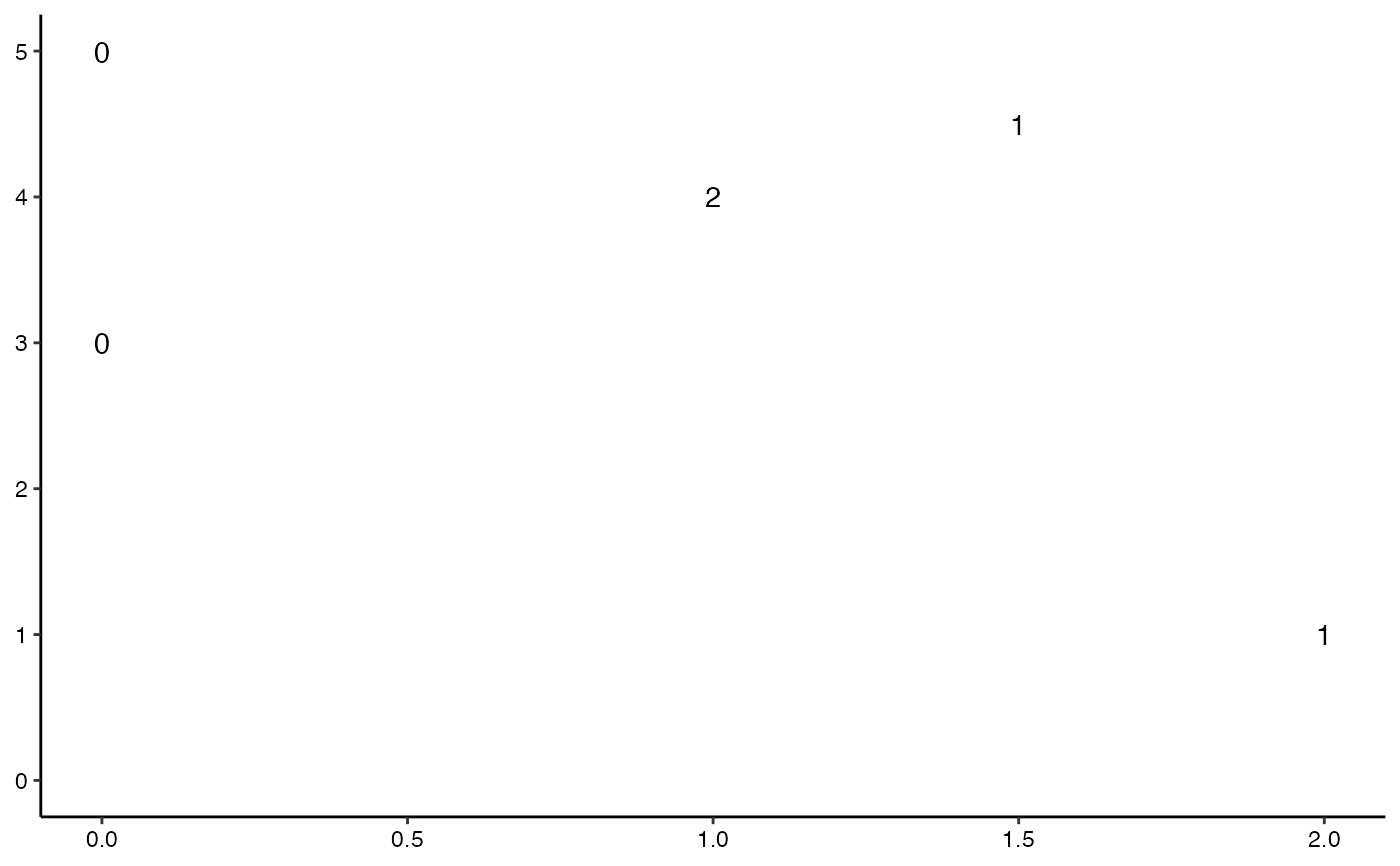
 # flat persistence diagram, coding dimension to numeral
ggplot(toy.data,
aes(start = birth, end = death, label = dim)) +
theme_persist() +
stat_persistence(diagram = "flat", geom = "text") +
lims(x = c(0, NA), y = c(0, NA))
# flat persistence diagram, coding dimension to numeral
ggplot(toy.data,
aes(start = birth, end = death, label = dim)) +
theme_persist() +
stat_persistence(diagram = "flat", geom = "text") +
lims(x = c(0, NA), y = c(0, NA))
 # flat persistence diagram, labeling by feature ID
ggplot(toy.data, aes(start = birth, end = death, colour = dim, shape = dim)) +
theme_persist() +
coord_equal() +
stat_persistence(
geom = "text",
aes(label = after_stat(id), alpha = after_stat(persistence)),
diagram = "flat", size = 3
) +
guides(alpha = "none")
# flat persistence diagram, labeling by feature ID
ggplot(toy.data, aes(start = birth, end = death, colour = dim, shape = dim)) +
theme_persist() +
coord_equal() +
stat_persistence(
geom = "text",
aes(label = after_stat(id), alpha = after_stat(persistence)),
diagram = "flat", size = 3
) +
guides(alpha = "none")
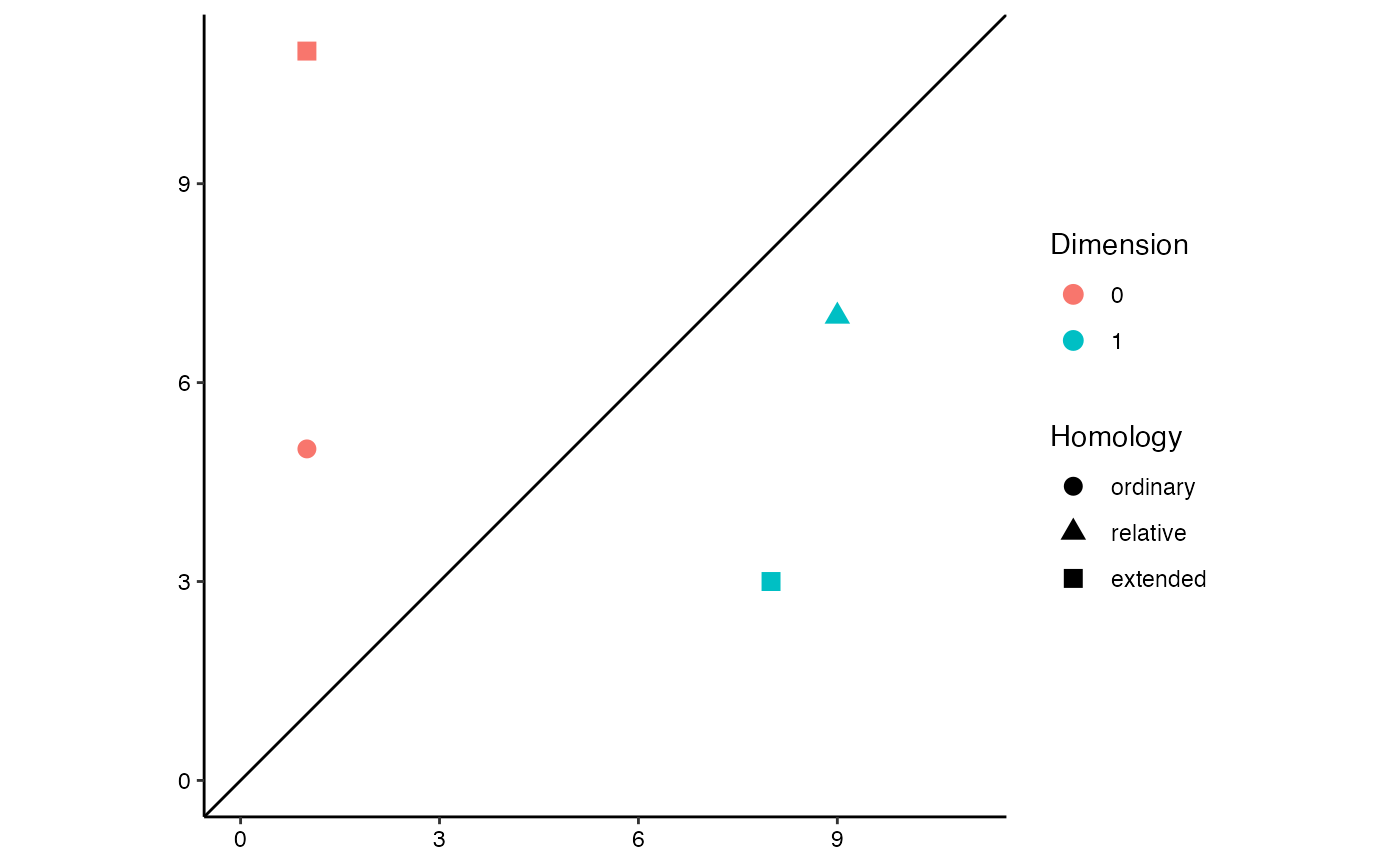
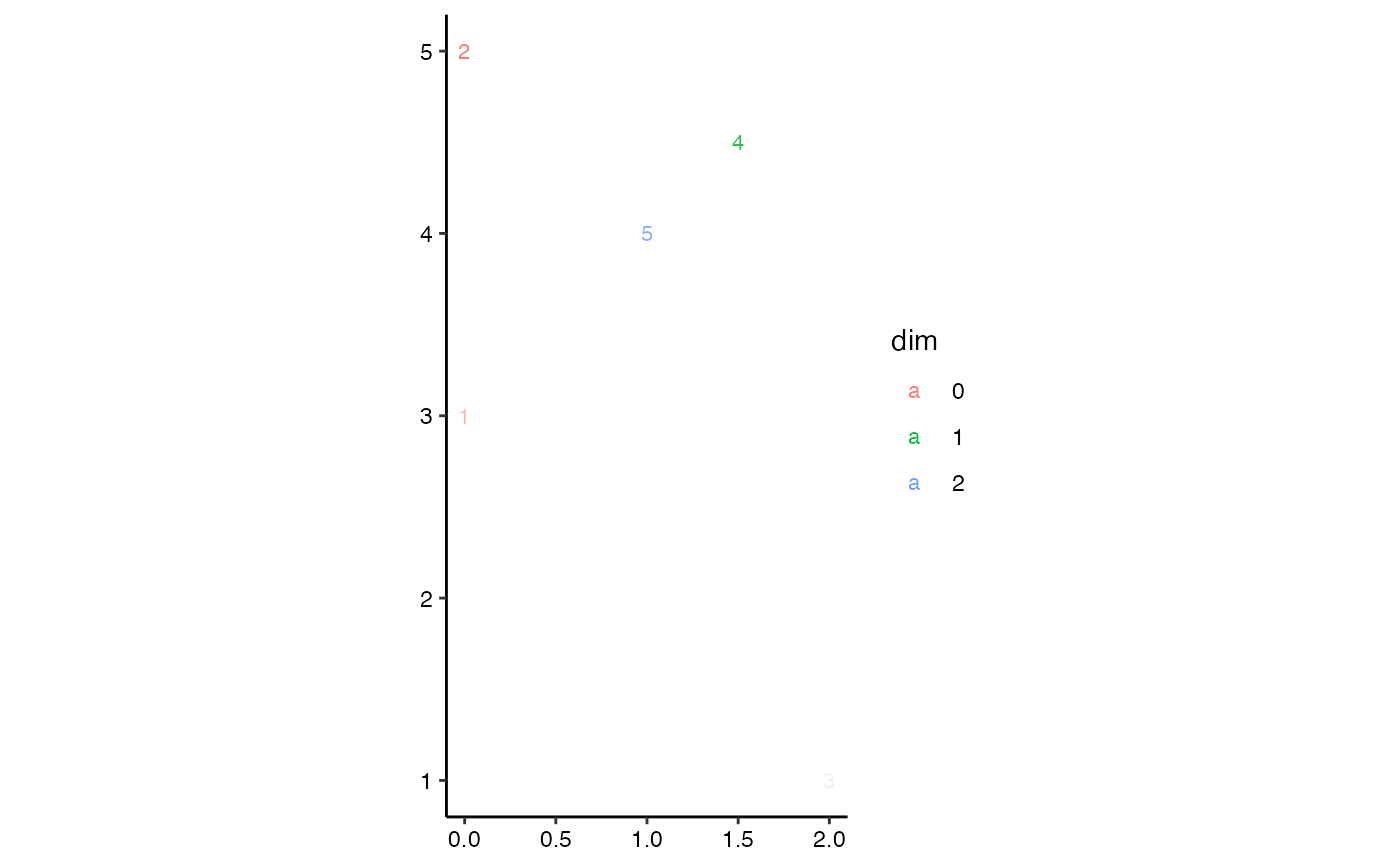 # toy extended persistence data, adapted from Carriere & Oudot (2015)
eph.data <- data.frame(
dimension = c(0L, 1L, 0L, 1L),
birth = c(1, -9, 1, 8),
death = c(5, -7, -11, -3)
)
# extended persistence diagram
ggplot(eph.data,
aes(start = birth, end = death, color = factor(dimension))) +
theme_persist() +
coord_equal() +
stat_persistence(aes(shape = after_stat(part)), size = 3) +
geom_abline(intercept = 0, slope = 1) +
lims(x = c(0, 11), y = c(0, 11)) +
labs(color = "Dimension", shape = "Homology")
# toy extended persistence data, adapted from Carriere & Oudot (2015)
eph.data <- data.frame(
dimension = c(0L, 1L, 0L, 1L),
birth = c(1, -9, 1, 8),
death = c(5, -7, -11, -3)
)
# extended persistence diagram
ggplot(eph.data,
aes(start = birth, end = death, color = factor(dimension))) +
theme_persist() +
coord_equal() +
stat_persistence(aes(shape = after_stat(part)), size = 3) +
geom_abline(intercept = 0, slope = 1) +
lims(x = c(0, 11), y = c(0, 11)) +
labs(color = "Dimension", shape = "Homology")
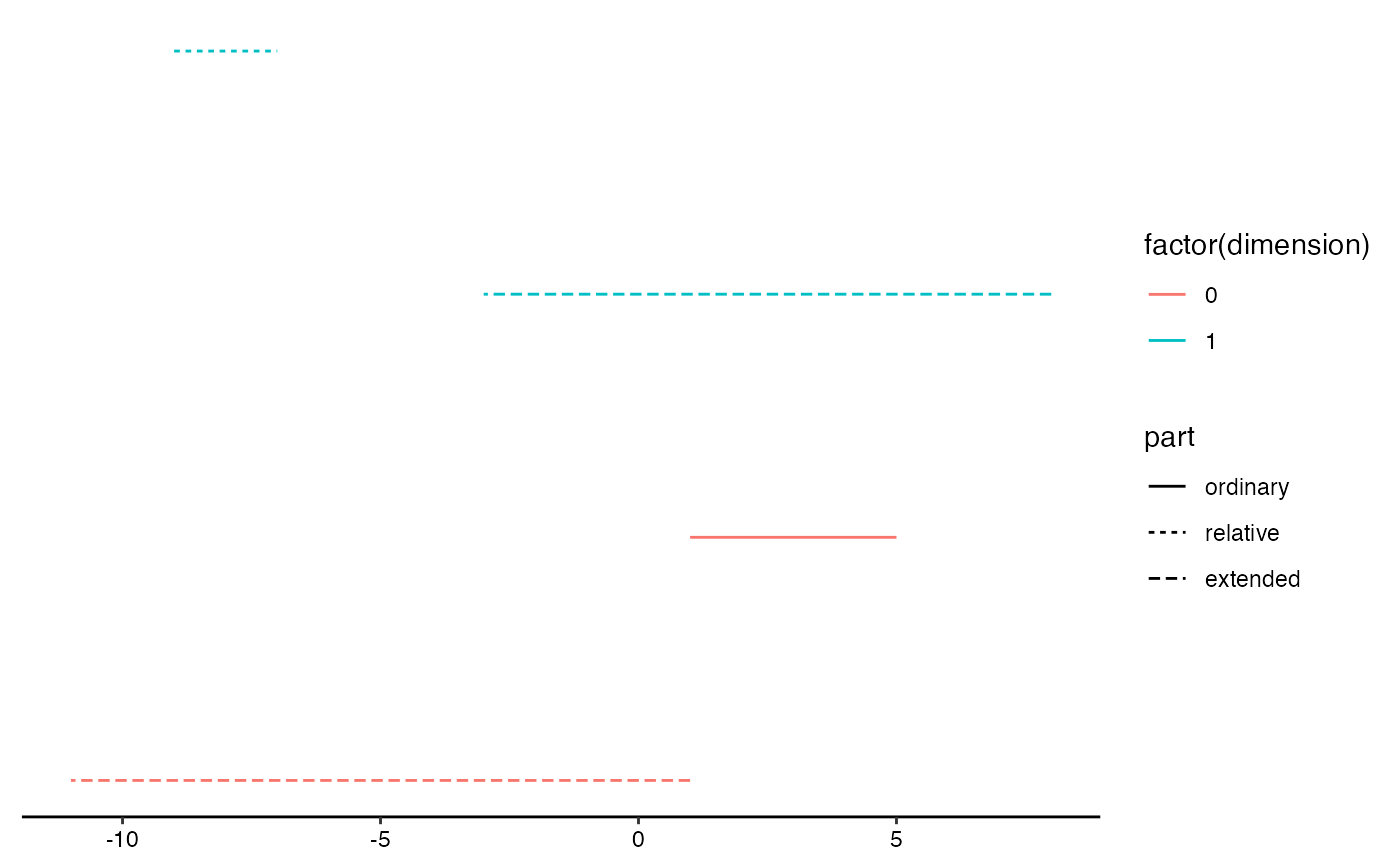
 # extended barcode
ggplot(eph.data,
aes(start = birth, end = death, color = factor(dimension))) +
theme_barcode() +
geom_barcode(aes(linetype = after_stat(part)))
# extended barcode
ggplot(eph.data,
aes(start = birth, end = death, color = factor(dimension))) +
theme_barcode() +
geom_barcode(aes(linetype = after_stat(part)))
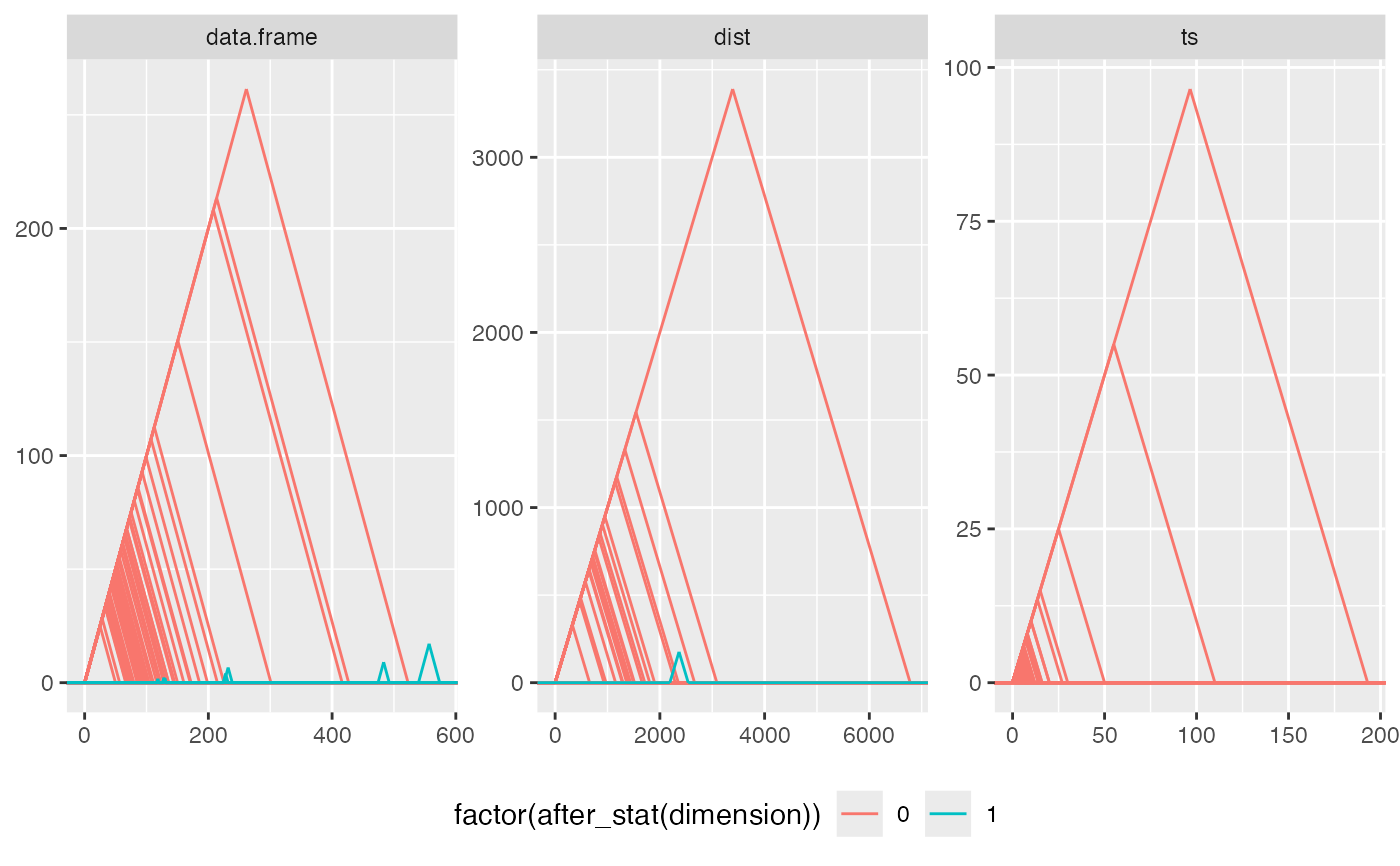
 # list-column of data sets to 'dataset' aesthetic
raw_data <- data.frame(obj = I(list(eurodist, 10*swiss, Nile)))
raw_data$class <- vapply(raw_data$obj, class, "")
if ("TDA" %in% rownames(utils::installed.packages())) {
# barcodes
ggplot(raw_data, aes(dataset = obj)) +
geom_barcode(aes(color = factor(after_stat(dimension))),
engine = "TDA") +
facet_wrap(facets = vars(class))
# persistence diagram
ggplot(raw_data, aes(dataset = obj)) +
stat_persistence(aes(color = factor(after_stat(dimension)), shape = class),
engine = "GUDHI")
# persistence landscape
ggplot(raw_data, aes(dataset = obj)) +
facet_wrap(facets = vars(class), scales = "free") +
stat_landscape(aes(color = factor(after_stat(dimension))),
engine = "Dionysus") +
theme(legend.position = "bottom")
}
#> Warning: Removed 3 rows containing non-finite outside the scale range
#> (`stat_landscape()`).
# list-column of data sets to 'dataset' aesthetic
raw_data <- data.frame(obj = I(list(eurodist, 10*swiss, Nile)))
raw_data$class <- vapply(raw_data$obj, class, "")
if ("TDA" %in% rownames(utils::installed.packages())) {
# barcodes
ggplot(raw_data, aes(dataset = obj)) +
geom_barcode(aes(color = factor(after_stat(dimension))),
engine = "TDA") +
facet_wrap(facets = vars(class))
# persistence diagram
ggplot(raw_data, aes(dataset = obj)) +
stat_persistence(aes(color = factor(after_stat(dimension)), shape = class),
engine = "GUDHI")
# persistence landscape
ggplot(raw_data, aes(dataset = obj)) +
facet_wrap(facets = vars(class), scales = "free") +
stat_landscape(aes(color = factor(after_stat(dimension))),
engine = "Dionysus") +
theme(legend.position = "bottom")
}
#> Warning: Removed 3 rows containing non-finite outside the scale range
#> (`stat_landscape()`).
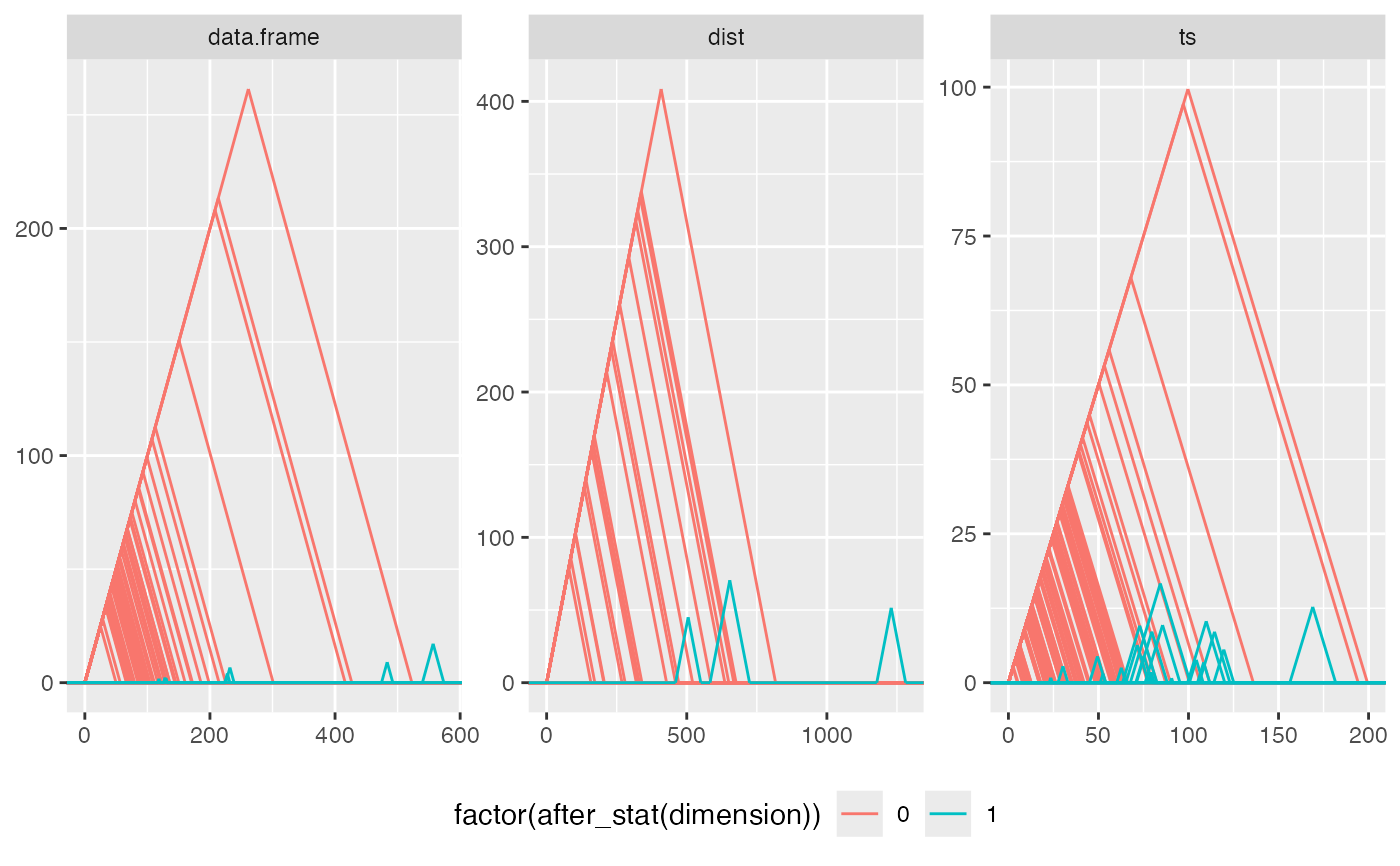
 if ("ripserr" %in% rownames(utils::installed.packages())) {
# exclude time series data if {ripserr} v0.1.1 is installed
if (utils::packageVersion("ripserr") == "0.1.1")
raw_data <- raw_data[c(1L, 2L), ]
# barcodes
ggplot(raw_data, aes(dataset = obj)) +
geom_barcode(aes(color = factor(after_stat(dimension))),
engine = "ripserr") +
facet_wrap(facets = vars(class))
# persistence diagram
ggplot(raw_data, aes(dataset = obj)) +
stat_persistence(aes(color = factor(after_stat(dimension)), shape = class),
engine = "ripserr")
# persistence landscape
ggplot(raw_data, aes(dataset = obj)) +
facet_wrap(facets = vars(class), scales = "free") +
stat_landscape(aes(color = factor(after_stat(dimension))),
engine = "ripserr") +
theme(legend.position = "bottom")
}
if ("ripserr" %in% rownames(utils::installed.packages())) {
# exclude time series data if {ripserr} v0.1.1 is installed
if (utils::packageVersion("ripserr") == "0.1.1")
raw_data <- raw_data[c(1L, 2L), ]
# barcodes
ggplot(raw_data, aes(dataset = obj)) +
geom_barcode(aes(color = factor(after_stat(dimension))),
engine = "ripserr") +
facet_wrap(facets = vars(class))
# persistence diagram
ggplot(raw_data, aes(dataset = obj)) +
stat_persistence(aes(color = factor(after_stat(dimension)), shape = class),
engine = "ripserr")
# persistence landscape
ggplot(raw_data, aes(dataset = obj)) +
facet_wrap(facets = vars(class), scales = "free") +
stat_landscape(aes(color = factor(after_stat(dimension))),
engine = "ripserr") +
theme(legend.position = "bottom")
}