The function step_pd_point_cloud() creates a specification
of a recipe step that will convert compatible data formats (distance
matrices, coordinate matrices, or time series) to 3-column matrix
representations of persistence diagram data. The input and output must be
list-columns.
Usage
step_pd_point_cloud(
recipe,
...,
role = NA_character_,
trained = FALSE,
filtration = "Rips",
max_hom_degree = 1L,
radius_max = NULL,
diameter_max = NULL,
field_order = 2L,
engine = NULL,
columns = NULL,
skip = FALSE,
id = rand_id("pd_point_cloud")
)Arguments
- recipe
A recipe object. The step will be added to the sequence of operations for this recipe.
- ...
One or more selector functions to choose variables for this step. See
selections()for more details.- role
For model terms created by this step, what analysis role should they be assigned? By default, the new columns created by this step from the original variables will be used as predictors in a model.
- trained
A logical to indicate if the quantities for preprocessing have been estimated.
- filtration
The type of filtration from which to compute persistent homology; one of
"Rips","Vietoris"(equivalent), or"alpha".- max_hom_degree, radius_max, diameter_max, field_order
Parameters passed to persistence engines.
- engine
The computational engine to use (see 'Details'). Reasonable defaults are chosen based on
filtration.- columns
A character string of the selected variable names. This field is a placeholder and will be populated once
prep()is used.- skip
A logical. Should the step be skipped when the recipe is baked by
bake()? While all operations are baked whenprep()is run, some operations may not be able to be conducted on new data (e.g. processing the outcome variable(s)). Care should be taken when usingskip = TRUEas it may affect the computations for subsequent operations.- id
A character string that is unique to this step to identify it.
Value
An updated version of recipe with the new step added to the
sequence of any existing operations.
Details
Persistent homology (PH) is a tool of algebraic topology to extract features from data whose persistence measures their robustness to scale. The computation relies on a sequence of maps between discrete topological spaces (usually a filtration comprising only inclusions) constructed from the data.
PH of Point Clouds
The PH of a point cloud arises from a simplicial filtration (usually Vietoris–Rips, Čech, or alpha) along an increasing distance threshold.
Ripser is a highly efficient implementation of PH on a point cloud (a
finite metric space) via the Vietoris–Rips filtration and is ported to R
through ripserr.
TDA calls the Dionysus, PHAT, and GUDHI libraries
to compute PH via Vietoris–Rips and alpha filtrations. The filtration
parameter controls the choice of filtration while the engine parameter
allows the user to manually select an implementation.
Both engines accept data sets in distance matrix, coordinate matrix, data frame, and time series formats.
The max_hom_degree argument determines the highest-dimensional features
to be calculated. Either diameter_max (preferred) or radius_max can be
used to bound the distance threshold along which PH is computed. The
field_order argument should be prime and will be the order of the field
of coefficients used in the computation. In most applications, only
max_hom_degree will be tuned, and to at most 3L.
Tuning Parameters
This step has 1 tuning parameter(s):
max_hom_degree: Maximum Homological Degree (type: integer, default: 1)
See also
Other topological feature extraction via persistent homology:
step_pd_degree(),
step_pd_raster()
Examples
roads <- data.frame(dist = I(list(eurodist, UScitiesD * 1.6)))
ph_rec <- recipe(~ ., data = roads) %>%
step_pd_point_cloud(dist, max_hom_degree = 1, filtration = "Rips")
ph_prep <- prep(ph_rec, training = roads)
ph_res <- bake(ph_prep, roads)
tidy(ph_rec, number = 1)
#> # A tibble: 1 × 3
#> terms value id
#> <chr> <dbl> <chr>
#> 1 dist NA pd_point_cloud_uKl5I
tidy(ph_prep, number = 1)
#> # A tibble: 1 × 3
#> terms value id
#> <chr> <dbl> <chr>
#> 1 dist NA pd_point_cloud_uKl5I
ops <- par(mfrow = c(1, 2), mar = c(2, 2, 0, 0) + 0.1)
for (i in seq(nrow(ph_res))) {
with(ph_res$dist[[i]], plot(
x = birth, y = death, pch = dimension + 1, col = dimension + 1,
xlab = NA, ylab = "", asp = 1
))
}
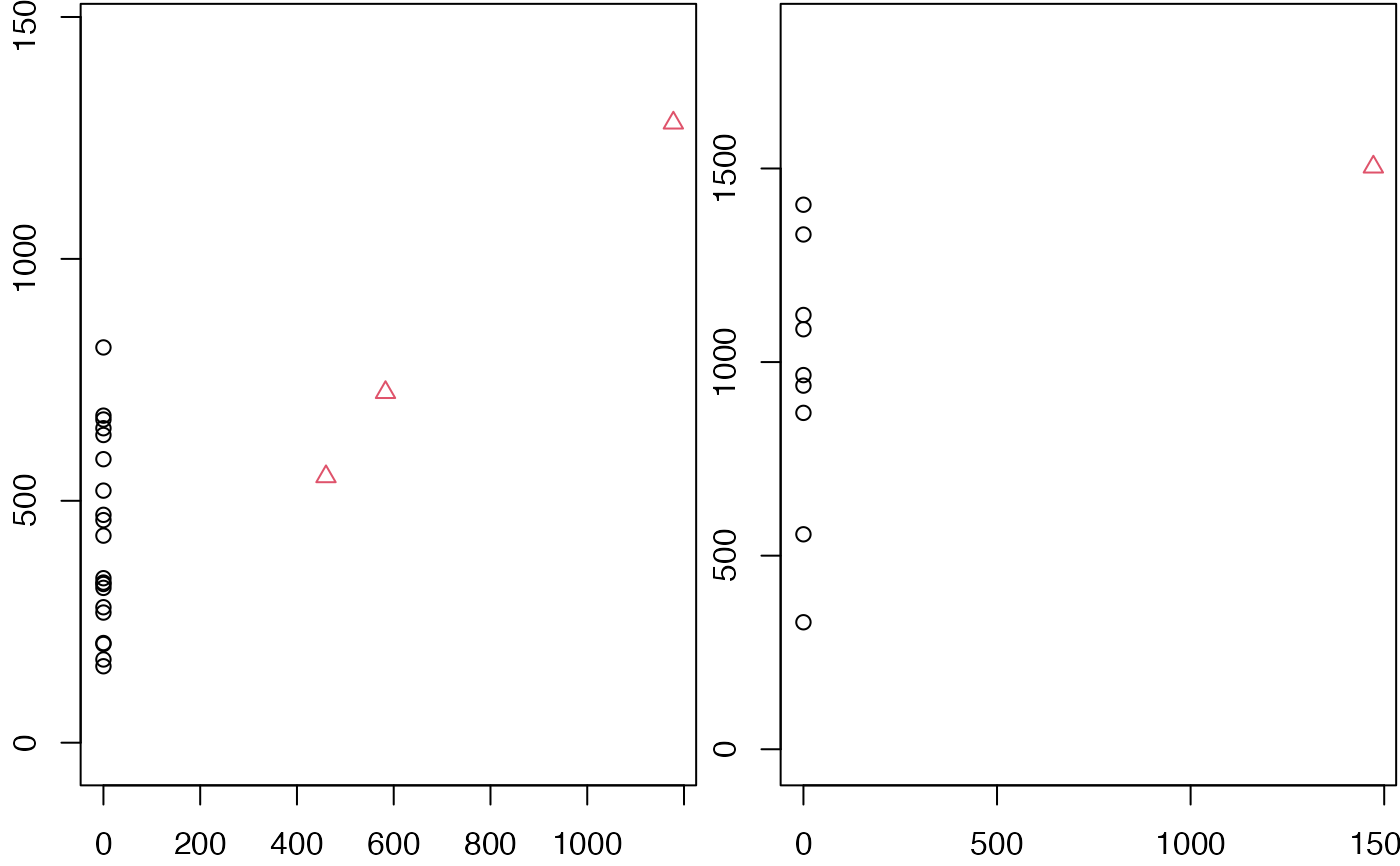 par(ops)
with_max <- recipe(~ ., data = roads) %>%
step_pd_point_cloud(dist, max_hom_degree = 1, diameter_max = 200)
with_max <- prep(with_max, training = roads)
bake(with_max, roads)
#> # A tibble: 2 × 1
#> dist
#> <list>
#> 1 <PHom [21 × 3]>
#> 2 <PHom [10 × 3]>
par(ops)
with_max <- recipe(~ ., data = roads) %>%
step_pd_point_cloud(dist, max_hom_degree = 1, diameter_max = 200)
with_max <- prep(with_max, training = roads)
bake(with_max, roads)
#> # A tibble: 2 × 1
#> dist
#> <list>
#> 1 <PHom [21 × 3]>
#> 2 <PHom [10 × 3]>