The function step_blur() creates a specification of a recipe
step that will induce Gaussian blur in numerical arrays. The input and
output must be list-columns.
Usage
step_blur(
recipe,
...,
role = NA_character_,
trained = FALSE,
xmin = 0,
xmax = 1,
blur_sigmas = NULL,
skip = FALSE,
id = rand_id("blur")
)Arguments
- recipe
A recipe object. The step will be added to the sequence of operations for this recipe.
- ...
One or more selector functions to choose variables for this step. See
selections()for more details.- role
For model terms created by this step, what analysis role should they be assigned? By default, the new columns created by this step from the original variables will be used as predictors in a model.
- trained
A logical to indicate if the quantities for preprocessing have been estimated.
- xmin, xmax, blur_sigmas
Parameters passed to
blur().- skip
A logical. Should the step be skipped when the recipe is baked by
bake()? While all operations are baked whenprep()is run, some operations may not be able to be conducted on new data (e.g. processing the outcome variable(s)). Care should be taken when usingskip = TRUEas it may affect the computations for subsequent operations.- id
A character string that is unique to this step to identify it.
Value
An updated version of recipe with the new step added to the
sequence of any existing operations.
Details
The gaussian blur step deploys blur(). See there for definitions
and references.
TODO: Explain the importance of blur for PH of image data.
Tuning Parameters
This step has 1 tuning parameter(s):
blur_sigmas: Gaussian Blur std. dev.s (type: double, default: NULL)
Examples
topos <- data.frame(pix = I(list(volcano)))
blur_rec <- recipe(~ ., data = topos) %>% step_blur(pix)
blur_prep <- prep(blur_rec, training = topos)
blur_res <- bake(blur_prep, topos)
tidy(blur_rec, number = 1)
#> # A tibble: 1 × 3
#> terms value id
#> <chr> <dbl> <chr>
#> 1 pix NA blur_FQJxb
tidy(blur_prep, number = 1)
#> # A tibble: 1 × 3
#> terms value id
#> <chr> <dbl> <chr>
#> 1 pix NA blur_FQJxb
with_sigmas <- recipe(~ ., data = topos) %>% step_blur(pix, blur_sigmas = 10)
with_sigmas <- bake(prep(with_sigmas, training = topos), topos)
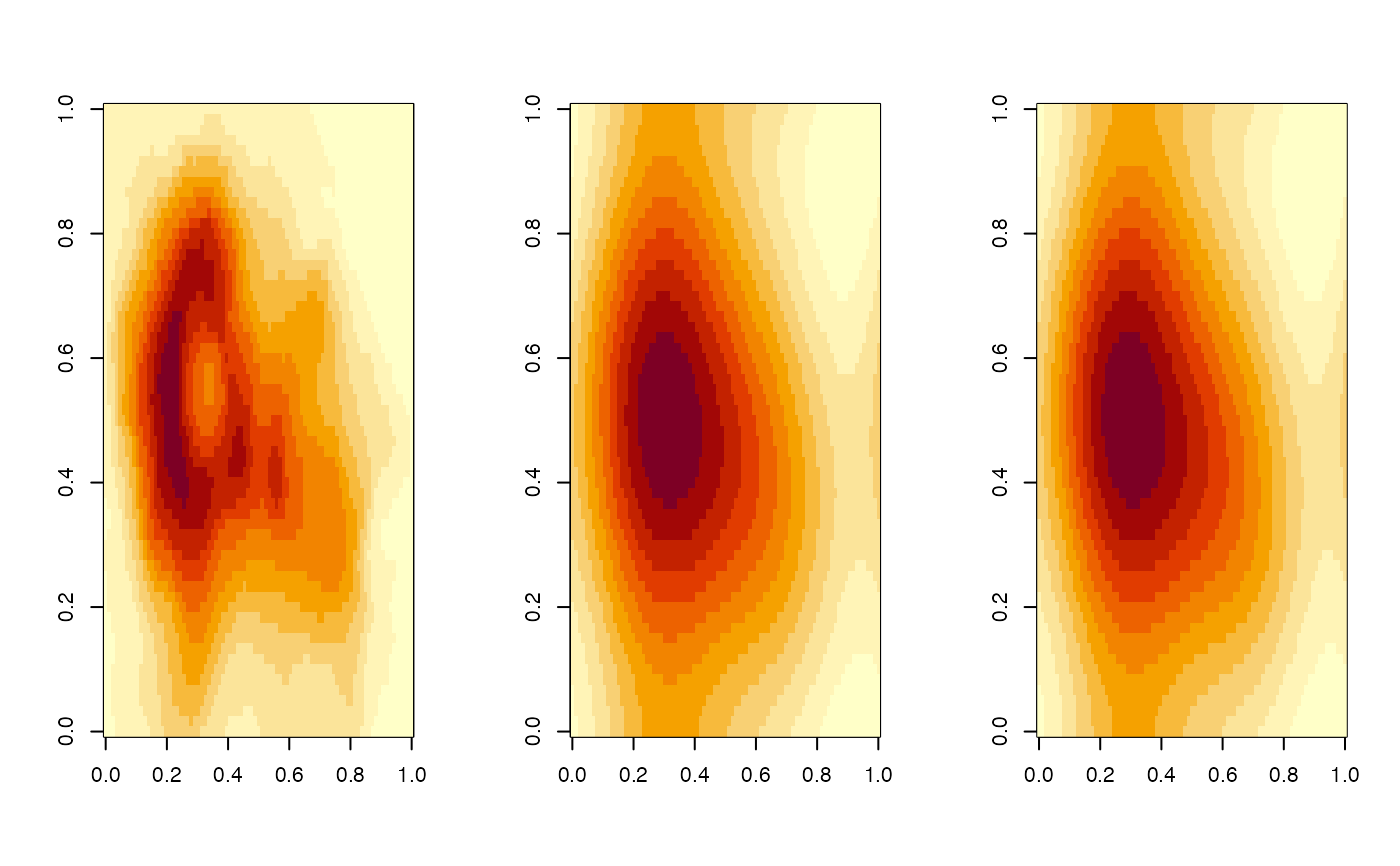
ops <- par(mfrow = c(1, 3))
image(topos$pix[[1]])
image(blur_res$pix[[1]])
image(with_sigmas$pix[[1]])
 par(ops)
par(ops)